Plant Sciences
Take your plant research to the next level!
ENVIRONMENT - PHENOTYPE - DATA
The Plant Sciences Facility (PlantS) operates 23 high quality and highly specialized plant growth chambers (phytotrons) and provides professional support to the outstanding “green research” at the Vienna BioCenter (VBC) and beyond.
The focus of the services provided is on environmental simulation, high-throughput (HT) plant phenotyping and subsequent image- and data analysis.
In 2017, PlantS initiated the Austrian Plant Phenotyping Network (APPN) with the goal to unite the Austrian plant phenotyping community and to support the ESFRI-EMPHASIS project.
SERVICES
High-throughput plant phenotyping
For objective, reproducible and high-throughput assessment of plant phenotypic traits we operate numerous phenotyping instruments for Arabidopsis and crop plants; shoot and root systems (see EQUIPMENT).
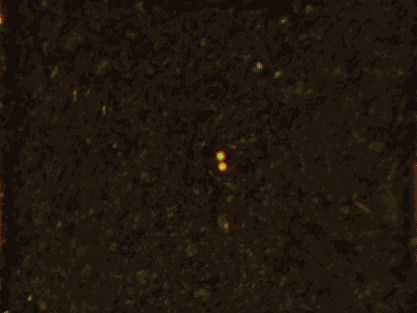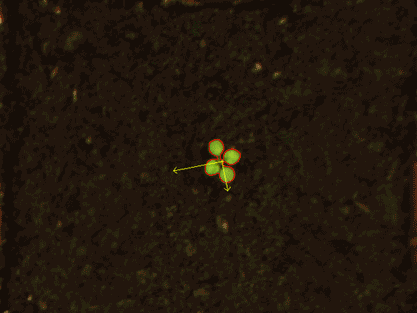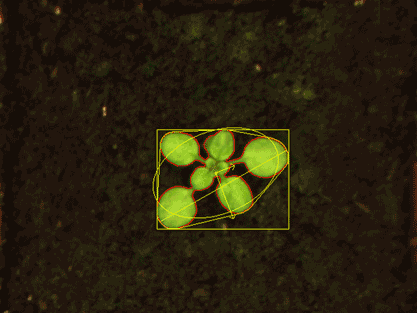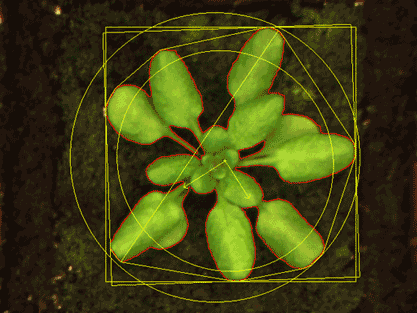
For the subsequent image analysis, we use classical image analysis approaches but also state-of-the-art deep learning pipelines. We offer a complete HT phenotyping service: from seeds to data analysis, statistics and data visualization.

Multi-sensor, high-throughput plant phenotyping platform - PHENOPlant
PHENOPlant is designed for non-invasive, morphometric and physiological hight-throughput plant phenotyping of mid-size crop plants as well as Arabidopsis and will be fully integrated into a state-of-the-art walk-in phytotron providing highly homogeneous plant growth conditions.
Furthermore, the platform facilitates precise environmental (live) simulations across different climate zones as well as controlled plant stress experiments. Cutting-edge sensors include multi-excitation PAM kinetic chlorophyll fluorescence, RGB, VNIR/SWIR hyperspectral, thermal and 3D.
Within the planned infrastructure, plants are transported on conveyor belts from the growth area to the imaging cabinets equipped with state-of-the-art sensors. The imaging units are physically seperated from the growth area (two individually controlled phytotrons) facilitating the parallelization of measurements via a sample loop and thus improving the throughput, flexibility and sensor utilization.
PHENOPlant 360° video from the point of view of a plant

The phenotyping platform PHENOTron is designed for HT top-view RGB imaging of small plants like Arabidopsis thaliana and is fully integrated into one of our high-tech phytotrons:

Environmental simulation
Several phytotrons are capable of providing exceptional environmental conditions, allowing precise environmental simulation across different climate zones and various abiotic stress conditions.
Simulated Environments:
- Cold stress (≥ -15°C) & heat stress (≤ +50°C)
- Water stress (drought & partial waterlogging)
- Variable light intensity & spectrum (LED-light: blue_405nm, blue_450nm, warm_white & red_660_730nm) incl. spectral simulation of sunrise and sunset
- CO2 level regulation
Custom Phenotyping and HT Image Analysis
The facility offers semi-automated phenotyping of various plant organs and growth stages, adapted to the needs of the customers:
PHENOBox: Side-view phenotyping of tall plants and agar-plates (seedlings).
PHENOScan: Agar-plate based screening of Arabidopsis roots.
HT Image Analysis: High-throughput image analysis service.
EQUIPMENT
With 23 high-quality, state-of-the-art and highly specialized plant growth chambers, PlantS can precisely control environmental conditions, reproducing abiotic plant stress conditions such as frost, drought and various spectral light and CO2 gas conditions. Furthermore, we can accurately simulate various global environmental conditions from a range of different climate zones.
PHENOPlant is designed for non-invasive, morphometric and physiological hight-throughput plant phenotyping of mid-size crop plants as well as Arabidopsis and will be fully integrated into a state-of-the-art walk-in phytotron providing highly homogeneous plant growth conditions.
The phenotyping platform (PHENOTron) is designed for top-view RGB imaging of small plants like Arabidopsis thaliana and is fully integrated into one of our high-tech phytotrons.
This particular phytotron facilitates frost stress experiments (> -15°C) and is equipped with a high-tech LED light with adjustable light spectrum.
A system for phenotyping of individual tall plants, e.g. model grass Brachypodium distachyon or maize. Coupled with PhenoPipe module for calculation of multi-dimensional distances from phenotyping data, we provide an affordable, automated, open source imaging and data processing solution that can be adapted to various phenotyping applications in plant biology and beyond.
Agar-plate based screening of Arabidopis roots
USER INFORMATION
In general, we offer four types of access to our shared research infrastructure:
- Research projects
- Full research services
- Trained user access | user labs
- Shared technology platform
Typically, these are set but not limited by the offered technology or instrument, and differ in the required user expertise, the usability of a technology, the user’s pre- and postprocessing input, and the underlying operational models.
Research project
Research projects are the equivalent of contract research organizations (CRO). The customer submits the starting material/sample and receives the ready-to-use data for publication. Hence, core facility members are often co-authoring and involved in the entire publication process.
Full research service
The user submits the sample, we perform a pre-defined workflow (incl. QC) and process the raw data. Data interpretation or contributions to publications cannot be offered in this service mode.
Trained user access | User labs
VBCF experts maintain an instrument park and train users to operate it. This requires a certain level of expertise, maturity of the offered technology, a hands-on attitude and reliability from the user.
Shared technology platform | Instrument park
These technologies require expert knowledge to run the offered instruments. Experts are hired by one of the research institutes on the Vienna BioCenter Campus. The experts maintain an instrument park, run the experiments and train other trainers. Machines can only be operated by experts.
The PlantS facility services can be accessed via research projects. The facility also provides full research service and trained user access.
All new facility users must attend a training session on general plant rules. Please contact Mrs. Anneliese Auer to book such a training. In case of technical problems, pest infestations, or other urgent issues please always write to: plants(at)vbcf.ac.at.
In general, the VBCF’s general cooperation conditions apply. In case of any questions on our services/equipment and for project requests please contact the Facility Head Jakub Jez.
We require acknowledgement of facility use in publications.
A simple statement is sufficient and can be placed in the Materials and Methods section or in the Acknowledgments section, depending on the journal format.
Suggested format:
The XXXXXX was performed by the Plant Sciences Facility at Vienna BioCenter Core Facilities (VBCF), member of the Vienna BioCenter (VBC), Austria.
In case of (co-)authorship:
The Vienna BioCenter Core Facilities (VBCF) Plant Sciences Facility acknowledges funding from the Austrian Federal Ministry of Education, Science & Research; and the City of Vienna.
SCIENTIFIC CONTRIBUTIONS
A bench-top Dark-Root device built with LEGO® bricks enables a non-invasive plant root development analysis in soil conditions mirroring nature. Dermendjiev G, Schnurer M, Stewart E, Nägele T, Marino G, Leister D, Thür A, Plott S, Jeż J, Ibl V. Frontiers in Plant Science, 2023, 14.
Metabolome plasticity in 241 Arabidopsis thaliana accessions reveals evolutionary cold adaptation processes. Weiszmann J, Walther D, Clauw P, Back G, Gunis J, Reichardt I, Koemeda S, Jez J, Nordborg M, Schwarzerova J, Pierides I, Nägele T, Weckwerth W. Plant Physiol. 2023, kiad298.
Many ways to TOPLESS - manipulation of plant auxin signalling by a cluster of fungal effectors. Bindics J, Khan M, Uhse S, Kogelmann B, Baggely L, Reumann D, Ingole KD, Stirnberg A, Rybecky A, Darino M, Navarrete F, Doehlemann G, Djamei A. New Phytol. 2022 236(4):1455-1470
Effector-mediated relocalization of a maize lipoxygenase protein triggers susceptibility to Ustilago maydis. Saado I, Chia KS, Betz R, Alcântara A, Pettkó-Szandtner A, Navarrete F, D'Auria JC, Kolomiets MV, Melzer M, Feussner I, Djamei A. Plant Cell. 34(7):2785-2805
TOPLESS promotes plant immunity by repressing auxin signaling and is targeted by the fungal effector Naked1. Navarrete F, Gallei M, Kornienko AE, Saado I, Khan M, Chia KS, Darino MA, Bindics J, Djamei A. Plant Commun. 2022 3(2):100269
Microtubules promote the non-cell autonomous action of microRNAs by inhibiting their cytoplasmic loading onto ARGONAUTE1 in Arabidopsis. Fan L, Zhang C, Gao B, Zhang Y, Stewart E, Jez J, Nakajima K, Chen X. Dev Cell. 2022 57(8):995-1008
Locally adaptive temperature response of vegetative growth in Arabidopsis thaliana. Clauw P, Kerdaffrec E, Gunis J, Reichardt-Gomez I, Nizhynska V, Koemeda S, Jez J, Nordborg M. Elife. 2022 11:e77913.
A CENH3 mutation promotes meiotic exit and restores fertility in SMG7-deficient Arabidopsis. Capitao C, Tanasa S, Fulnecek J, Raxwal VK, Akimcheva S, Bulankova P, Mikulkova P, Crhak Khaitova L, Kalidass M, Lermontova I, Mittelsten Scheid O, Riha K. PLoS Genet. 2021 17(9):e1009779.
A hypomorphic allele of telomerase uncovers the minimal functional length of telomeres in Arabidopsis. Watson JM, Trieb J, Troestl M, Renfrew K, Mandakova T, Fulnecek J, Shippen DE, Riha K. Genetics. 2021 219(2):iyab126.
Repression of CHROMOMETHYLASE 3 prevents epigenetic collateral damage in Arabidopsis. Papareddy RK, Páldi K, Smolka AD, Hüther P, Becker C, Nodine MD. Elife. 2021 10:e69396.
Comparative transcriptomic analysis reveals conserved programmes underpinning organogenesis and reproduction in land plants. Julca I, Ferrari C, Flores-Tornero M, Proost S, Lindner AC, Hackenberg D, Steinbachová L, Michaelidis C, Gomes Pereira S, Misra CS, Kawashima T, Borg M, Berger F, Goldberg J, Johnson M, Honys D, Twell D, Sprunck S, Dresselhaus T, Becker JD, Mutwil M. Nat Plants. 2021 7(8):1143-1159.
Gene expression variation in Arabidopsis embryos at single-nucleus resolution. Kao P, Schon MA, Mosiolek M, Enugutti B, Nodine MD. Development. 2021 148(13):dev199589.
Crosstalk between H2A variant-specific modifications impacts vital cell functions. Schmücker A, Lei B, Lorković ZJ, Capella M, Braun S, Bourguet P, Mathieu O, Mechtler K, Berger F. PLoS Genet. 2021 17(6):e1009601.
Application of expansion microscopy on developing Arabidopsis seeds. Kao P, Nodine MD. Methods Cell Biol. 2021;161:181-195.
Epigenetic reprogramming rewires transcription during the alternation of generations in Arabidopsis. Borg M, Papareddy RK, Dombey R, Axelsson E, Nodine MD, Twell D, Berger F. Elife. 2021 10:e61894.
Polyploidy-associated paramutation in Arabidopsis is determined by small RNAs, temperature, and allele structure. Bente H, Foerster AM, Lettner N, Mittelsten Scheid O. PLoS Genet. 2021 17(3):e1009444.
The chromatin remodeler DDM1 prevents transposon mobility through deposition of histone variant H2A.W. Osakabe A, Jamge B, Axelsson E, Montgomery SA, Akimcheva S, Kuehn AL, Pisupati R, Lorković ZJ, Yelagandula R, Kakutani T, Berger F. Nat Cell Biol. 2021 23(4):391-400.
Signatures of antagonistic pleiotropy in a bacterial flagellin epitope. Parys K, Colaianni NR, Lee HS, Hohmann U, Edelbacher N, Trgovcevic A, Blahovska Z, Lee D, Mechtler A, Muhari-Portik Z, Madalinski M, Schandry N, Rodríguez-Arévalo I, Becker C, Sonnleitner E, Korte A, Bläsi U, Geldner N, Hothorn M, Jones CD, Dangl JL, Belkhadir Y. Cell Host Microbe. 2021 29(4):620-634.e9.
Autophagy mediates temporary reprogramming and dedifferentiation in plant somatic cells Rodriguez E, Chevalier J, Olsen J, Ansbøl J, Kapousidou V, Zuo Z, Svenning S, Loefke C, Koemeda S, Serrano Drozdowskyj P, Jez J, Durnberger G, Kuenzl F, Schutzbier M, Mechtler K, Nagel Ebstrup E, Lolle S, Dagdas Y, Petersen M. EMBO J (2020).
Chromatin Organization in Early Land Plants Reveals an Ancestral Association between H3K27me3, Transposons, and Constitutive Heterochromatin. Montgomery SA et al. [2020], Current Biology 30:4, 573-588.e7
The evolution and functional divergence of the histone H2B family in plants. Jiang D, Borg M, Lorković ZJ, Montgomery SA, Osakabe A, Yelagandula R, Axelsson E, Berger F. [2020], PLoS Genet 16(7): e1008964.
Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Borg M et al. [2020], Nature Cell Biology 22: 621–629
ARADEEPOPSIS, an Automated Workflow for Top-View Plant Phenomics using Semantic Segmentation of Leaf States. Hüther P, Schandry N, Jandrasits K, Bezrukov I, Becker C [2020], Plant Cell doi.org/10.1105/tpc.20.00318
Versatile in vitro assay to recognize Cas9-induced mutations. Bente H, Mittelsten Scheid O, Donà M. [2020] Plant Direct. 4: 1– 13.
Chromatin regulates expression of small RNAs to help maintain transposon methylome homeostasis in Arabidopsis. Papareddy RK, Páldi K, Paulraj S, Kao P, Lutzmayer S, Nodine MD [2020] Genome Biology 21, 251.
Arabidopsis shoot stem cells display dynamic transcription and DNA methylation patterns. Gutzat R, Rembart K, Nussbaumer T, Hofmann F, Pisupati R, Bradamante G, Daubel N, Gaidora A, Lettner N, Donà M, Nordborg M, Nodine M, Mittelsten Scheid O. [2020] EMBO J. 39(20):e103667.
A high-throughput screening method to identify proteins involved in unfolded protein response of the endoplasmic reticulum in plants. Alcântara A, Seitner D, Navarrete F, Djamei A. [2020] Plant Methods. 16:4.
Ribosome assembly factor Adenylate Kinase 6 maintains cell proliferation and cell size homeostasis during root growth.Slovak R, Setzer C, Roiuk M, Bertels J, Göschl C, Jandrasits K, Beemster GTS, Busch W. [2020] New Phytol. 225(5):2064-2076.
A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress.Stephani M, Picchianti L, Gajic A, Beveridge R, Skarwan E, Sanchez de Medina Hernandez V, Mohseni A, Clavel M, Zeng Y, Naumann C, Matuszkiewicz M, Turco E, Loefke C, Li B, Dürnberger G, Schutzbier M, Chen HT, Abdrakhmanov A, Savova A, Chia KS, Djamei A, Schaffner I, Abel S, Jiang L, Mechtler K, Ikeda F, Martens S, Clausen T, Dagdas Y. [2020] Elife 9:e58396.
The'PhenoBox', a flexible, automated, open-source plant phenotyping solution
Czedik-Eysenberg A, Seitner S, Güldener U, Koemeda S, Jez J, Colombini M, Djamei A. The New Phytologist, 2018.